 Seasonal Coronavirus Epidemiology & Evolution
Seasonal Coronavirus Epidemiology & Evolution
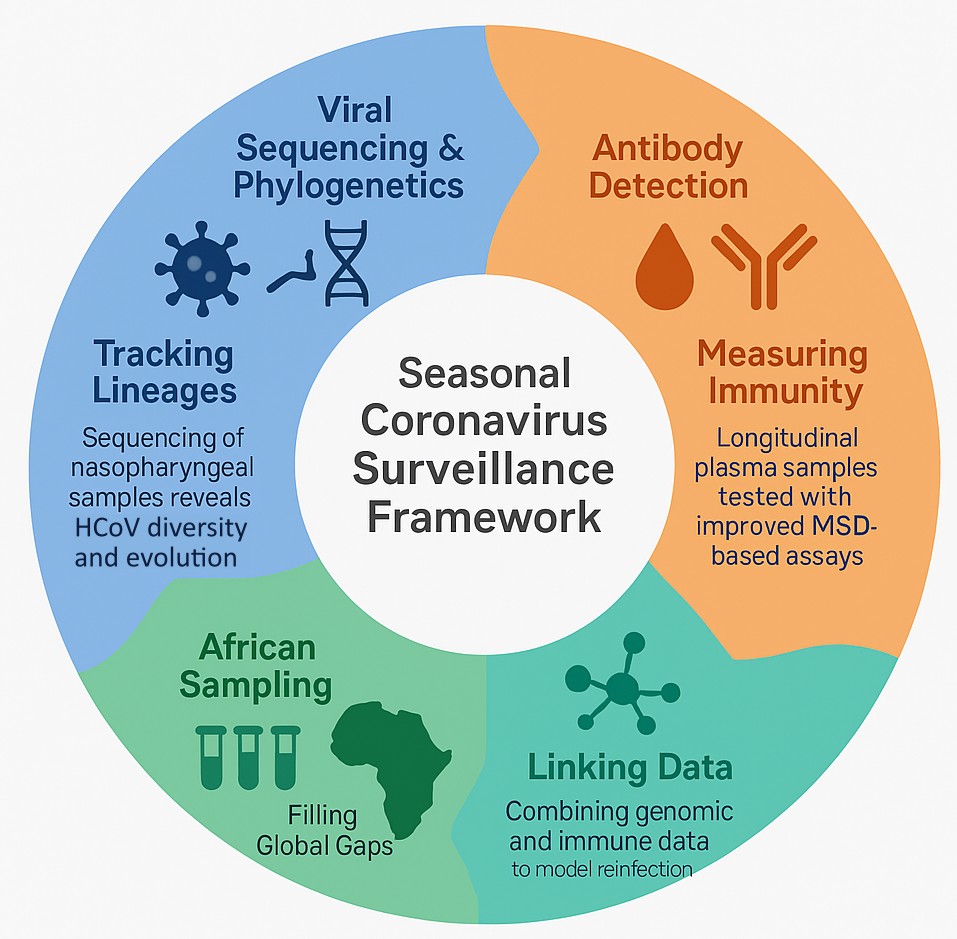
Our research on seasonal human coronaviruses (HCoVs) seeks to understand how these widespread viruses evolve, circulate, and interact with human immunity over time. We are particularly interested in how reinfections occur, the roles of immune waning and antigenic change, and the potential for under-sampled regions to reshape our understanding of HCoV diversity. To address these questions, we are building real-time phylogenies of HCoVs, exploring recombination events and genomic evolution, and integrating this with serological data to assess patterns of past infection. One of our goals is to improve pan-coronavirus sequencing protocols and to identify conserved regions - especially within the nucleocapsid (N) protein - that can be used to develop more accurate serological assays with reduced cross-reactivity across HCoV species.
In collaboration with Prof Katharina Röltgen’s VIM group, we are testing nasopharyngeal (NP) swabs and longitudinal plasma samples from Africa to study both active and past HCoV infections. Sequencing of NP samples helps us characterize the circulating viral lineages in the region, while electrochemiluminescence-based serological assays allow us to track antibody responses over time. By combining these approaches, we aim to uncover how immunity to HCoVs varies across geographies and age groups, and how this shapes viral evolution. These efforts are not only deepening our understanding of HCoVs, but also creating tools and frameworks that could be rapidly adapted in future coronavirus outbreaks.
- Check out some of our Nextstrain HCoV builds Coming soon