Enterovirus D68 now on Nextclade!
Check out the new Nextclade dataset for Enterovirus D68!
We’re excited to announce the launch of the first Nextclade dataset for Enterovirus D68 (EV-D68) - a major step toward easier, faster, and more consistent enterovirus surveillance. With EV-D68 cases already rising this season, clinicians and researchers can now use Nextclade to rapidly analyze their viral sequences, check sequence quality, identify clades, and compare results to a global phylogenetic tree - all within minutes in your web browser and without specialized software. Created together with the European Non-Polio Enterovirus Network (ENPEN), this first Enterovirus dataset in Nextclade lays the groundwork for future Enterovirus datasets and coordinated surveillance efforts.
You can explore the new Enterovirus D68 dataset here!
This dataset was created by Nadia Neuner-Jehle, Alejandra González Sánchez, and Emma B. Hodcroft.
Dataset details
The EV-D68 dataset is based on curated, high-quality whole genome sequences from public databases. It supports:
- Clade assignments (A1–A2, B1–B3, and C)
- Mutation calling relative to the reference
- Quality control (QC) metrics
- Gene annotations across the genome
Using whole-genome data allows reliable clade assignments even when VP1 is missing - something that wasn’t possible with earlier approaches.
Nextclade runs completely within your browser - no data is uploaded or sent anywhere, making it a completely private way to examine your sequences.
How to use the EV-D68 dataset in Nextclade
-
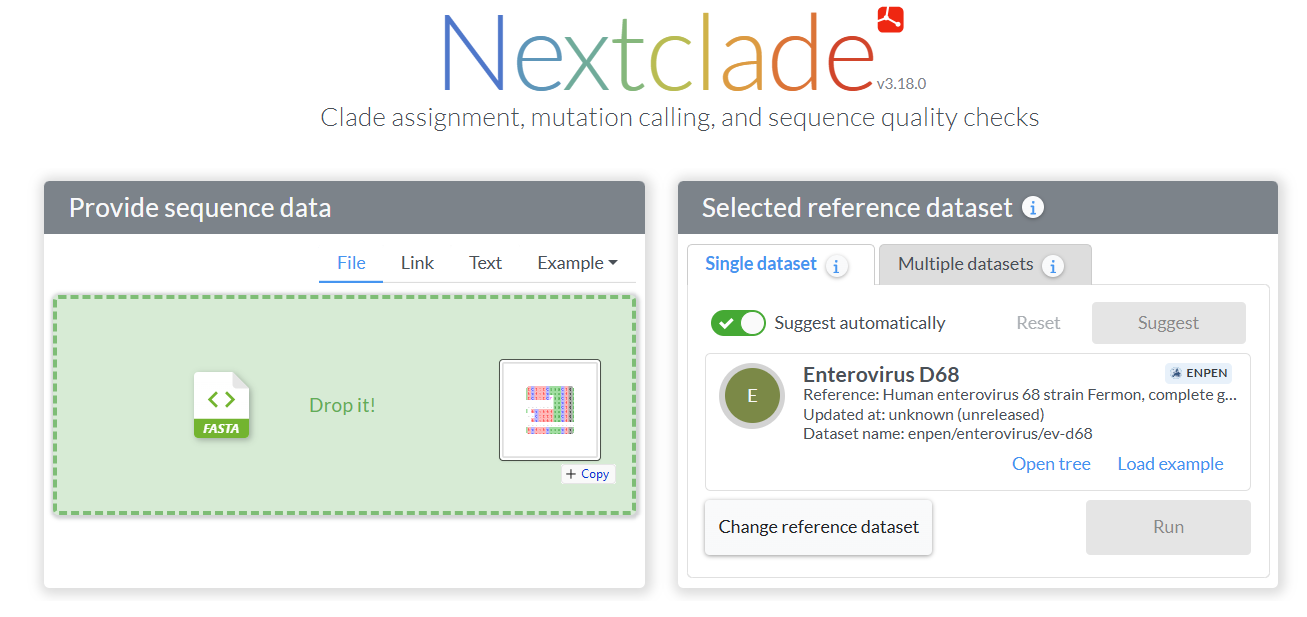
Add your sequences and run the analysis
Go to nextclade.org, choose the Enterovirus D68 dataset, and either drag and drop your FASTA file or load an example. Click “Run” to start the analysis. (If “Suggest automatically” is turned on, you can also add your FASTA file and let it detect the EV-D68 dataset automatically.)
Nextclade will align your sequences, assess quality, and assign clades automatically. All processing happens directly in your web browser.
-
Explore your results
Once processed, you’ll be taken to the Results section. Here, you can view sequence names (1), quality control (QC) metrics (2), clade assignments (3), and details such as the number of mutations (4), coverage, and gaps relative to the reference.
The “Genetic feature” panel, located to the right of the screen, displays either the entire genome (when “Nucleotide sequence” is selected, as shown in the image) or a single gene, such as VP1. The colored lines represent mutations relative to the reference sequence. Hovering over the lines provides more details on the mutations. If viewing the whole genome, this is shown in nucleotides; if viewing a single gene, it is shown in amino acids. Grayed-out regions indicate missing data. Even if only partial regions are provided (e.g., 3D-pol), clade assignment is usually still accurate in EV-D68, due to the low recombination rate.
References
You can change the reference against which your results are compared using the drop-down menu “Relative to” in the top-right corner:
- Reference: The default reference is Fermon, and all sequences are aligned against it.
- Parent: The closest parental node in the tree.
- Clade founder: The most recent common ancestor of all sequences sharing the same clade (e.g. B3 clade founder), inferred from the dataset.
- Static Inferred Ancestor: A stable sequence close to the root that we have derived from an inferred ancestral node and that will not change in the future. We provide this as a reference because it is equidistant from all sequences in the tree, and more closely related to currently circulating sequences than Fermon. You can view this sequence here.
- Tree root: The dataset root, which may shift slightly between dataset releases.
Clade Assignments
Clade assignments are based on the global diversity of Enterovirus D68. Current clades include A (A1–A2), B (B1–B3), and C.
Clade A2 is sometimes referred to as clade D; therefore, this dataset uses the designation “A2/D”.
The label “pre-ABC” indicates very old basal lineages that are no longer circulating. If your sequence is labeled as “pre-ABC” or “unassigned”, it may suggest sequencing or assembly issues rather than a true novel lineage — please check carefully!
The color of each row reflects your sequence’s QC status: 👉🏻
- Red: low quality
- Yellow: moderate quality
- White: high-quality sequence
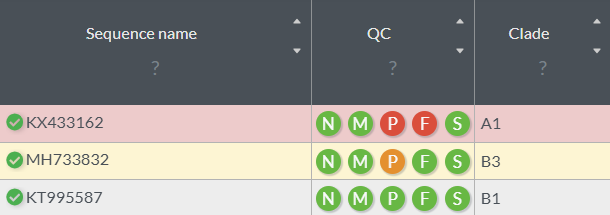

👈🏻 You can check which QC metric triggered a warning. For example, a red “P” indicates an unusually high number of private mutations, and the “F” is highlighted because the frame shift is not expected.
See the Nextclade guide for more details on QC metrics and result interpretation.
-
View your sequences in the phylogenetic tree
In the Tree view (button on the top-left), your sequences appear within the global reference phylogeny. By default, the sequence you provided will be shown as larger circles, and the branches they are connected to, by larger lines (1). The sequences in the reference dataset appear as the thinner lines (2). To view both your sequences and the reference tree together, toggle the 👁️ ‘eye’ icon (hide) at the top left of the tree (3), or remove the filter altogether with the 🗑️ ‘trash’ icon (remove).
Just as on a Nextstrain tree, you can color branches by metadata such as “QC status” or “Clade” (4). Some color schemes, such as Country, will apply only to the reference tree. You can also apply other filters to the data, or view the tree in different formats (such as radial view).
The reference tree provides a curated, down-sampled view of global EV-D68 diversity and we aim to update it at least once a year to reflect new data.
-
Learn more about the dataset
Clicking “Dataset” at the top leads to a detailed description of the its scope. This description includes important information about the current version of the dataset, which could be useful for recreating analyses. The description includes the reference sequence, dataset maintainers, dataset history, features, subgenogroup/clade definitions, reference types, and links to GitHub, documentation, and contact details.
-
Export and share your results
Use “Export” to download alignments, results tables, and phylogenetic trees in various formats such as CSV/TSV/JSON files containing mutation and clade information, as well as tree and annotation files. These files are ideal for sharing or further analysis.
Looking ahead
Work is already underway to expand Nextclade to other Enteroviruses, including EV-A71, CVA10, and CVA16.
Some features we hope to include in the future are:
- A recombination QC flag to identify likely recombinant genomes
- Gene-specific clade assignments (e.g., for VP1 and 3D)
These additions will further enhance the ability to quickly and easily obtain clade assignments and quality control information about more Enteroviruses.
While Nextclade provides a fast and accessible overview, and reliable QC and general clade assignment, it is not a replacement for full phylogenetic analyses. To conduct in-depth evolutionary and recombination studies, explore our Nextstrain Enterovirus trees at nextstrain.org/groups/hodcroftlab, which include VP1, whole-genome, and tanglegram views. Or - check out the Nextstrain tutorial to start setting up your own local build!
We encourage clinicians, researchers, and surveillance teams to explore the new EV-D68 dataset and share their feedback on GitHub or via email. Your input helps improve these tools and supports coordinated Enterovirus surveillance worldwide.
If you use this dataset in a publication, please cite Neuner-Jehle, N., González Sánchez, A., Hodcroft, E. B., & ENPEN. (2025) and Aksamentov et al. (2021). Citing this work helps us demonstrate its impact and secure resources for continued maintenance!
Links:


